Protein Stability Prediction Tool
You will surely end up with something really amazing! Find Buggy Plans barracuda for building that perfect Barracuda buggy you have long been waiting to build. Dune buggy blueprints free. Are you worried that your budget won’t make it? Or personalized your by adding arts like Kite buggy drawings that suits your taste! You can always opt for second hand buggy parts for building your own off road buggy these second hands comes in handy if you have a budget to think about!
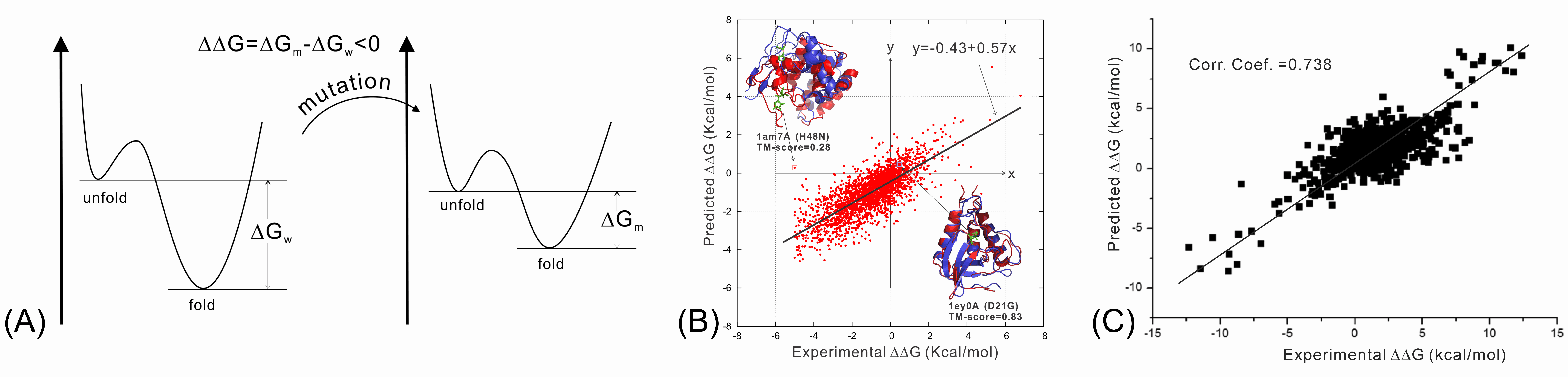
STRUM is a method for predicting the fold stability change (ΔΔG) of protein molecules upon single-point nsSNP mutations. STRUM adopts a gradient boosting regression approch to train the Gibbs free-energy changes on a variety of features at different levels of sequence and structure properties.
Mutation of a single amino acid residue can cause changes in a protein, which could then lead to a loss of protein function. Predicting the protein stability.

The unique characteristics of STRUM is the combination of sequence profiles with low-resolution structure models from protein structure prediction, which helps enhance the robustness and accuracy of the method and make it applicable to various protein seqences, including those without experimental structures (>> ). [] [] [] [] Please copy and paste your data below (either [10,1500] residues sequence in or experimental structure in is acceptable) Or upload the sequence/stucture file: Mode I: Single-point mutations Mode II: Systematic Email: (Mandatory, where results will be sent to. Only academic emails will be accepted.) ID: (Optional, your given name to this protein) STRUM Resource: References: • Lijun Quan, Qiang Lv, Yang Zhang. STRUM: Structure-based stability change prediction upon single-point mutation, Bioinformatics, 32: 2911-19 (2016) (download or ) (734) 647-1549 100 Washtenaw Avenue, Ann Arbor, MI.